Justin Angevaare

About me
I am a Data Scientist at Wellington-Dufferin-Guelph Public Health. I have an MSc (2014) and PhD (2020) in Applied Statistics from the University of Guelph.
In my research I navigate the challenges in the statistical modelling of complex population processes. Applications in public health and population ecology are of particular interest to me. This involves me in the study of topics in computational statistics, spatiotemporal models, and individual and agent based models. Much of my recent work has been implemented in open-source software packages.
Current software projects
Pathogen.jl (2015-present)Pathogen.jl is an implementation of some of my PhD research into transmission network individual level models of infectious disease, and phylodynamic models in Julia. It seeks to provide accessible and flexible tools for the simulation, description, visualization, and Bayesian inference of such models. This software provides methods for imputation of event times and transmission networks using data augmentation

SubstitutionModels.jl is a package that I've contributed to the BioJulia organization. At this time, it provides performant utilities for dealing with nucleic acid substitution models using StaticArrays.jl and Julia's multiple dispatch. These models are used for genetic distance calculations, phylogenetic tree construction, genetic sequence simulation, etc.
PhyloTrees.jl (2016-present)PhyloTrees.jl provides a representation of rooted phylogenetic trees in Julia. My eventual goal is to implement Bayesian phylogenetic tree inference using MCMC in Julia with this package. For the time being it has been used in the implementation of phylodynamic models as part of my PhD research.
GeneticBitArrays.jl uses Julia's BitArrays to represent RNA and DNA sequences. This representation is space efficient and highly interpretable. BitArrays also work great with linear algebra in Julia - such as may be used for phylogenetic inference, and simulation of genetic sequences.
PhyloModels.jl builds on my work with GeneticBitArrays.jl, PhyloTrees.jl, SubstitutionModels.jl. It implements Felsenstein's Pruning Algorithm for loglikelihood calculation of phylogenetic trees with aligned genetic sequences at their tips. A Dict with Int64 node IDs as keys and GeneticSeq as values is used for input for the loglikelihood calculation, as well as the output from simulation of genetic sequences.
Articles
Published- Angevaare, J., Feng, Z. and Deardon, R. (2021) Inference of latent event times and transmission networks in individual level infectious disease models. Spatial and Spatiotemporal Epidemiology. 10.1016/j.sste.2021.100410 | preprint version
- Angevaare, J., Feng, Z., Deardon, R. (2022) Pathogen.jl: Infectious disease transmission network modelling with Julia. Journal of Statistical Software. 10.18637/jss.v104.i04
Proceedings
- Rose, D., Edwards, B., Kett, R., Gillis, D., Angevaare, J. (2017) Exploring Anthropogenic Activities and Management Decisions Using a Novel Environmental Agent Based Model. IEEE International Humanitarian Technology Conference, Toronto, ON
Presentations
- Angevaare, J.✦, Feng, Z., Deardon, R. (2017) Simulation and inference of phylodynamic individual level models. Epidemics 6 International Conference on Infectious Disease Dynamics. Sitges, Spain (contributed oral presentation)
- Angevaare, J.✦, Feng, Z., Deardon, R. (2016) Phylodynamic individual level models: strategies for simulation and inference. Southwestern Ontario Graduate Mathematics and Statistics Conference, Guelph, ON (contributed poster presentation)
- Angevaare, J.✦, Feng, Z., Deardon, R. (2016) Phylodynamic individual level models: strategies for simulation and inference. Joint Statistical Meetings, Chicago, IL (contributed poster presentation)
- Angevaare, J.✦, Feng, Z., Deardon, R. (2016) Phylodynamic individual level models: strategies for simulation and inference. Annual Meeting of the Statistical Society of Canada, Brock University, St. Catharines, ON (contributed poster presentation)
- Angevaare, J.✦, Feng, Z., Deardon, R. (2016) A Phylodynamic extension to individual level models. Canadian Association of Veterinary Epidemiology and Preventive Medicine Conference, University of Guelph, Guelph, ON (contributed poster presentation)
- Rose, D.✦, Kett, R., Yodzis, M., Angevaare, J., Gillis, D. (2015) A combined agent and stage structured model to investigate anthropogenic activities on a wild fish population. College of Physical and Engineering Science Undergraduate Poster Session, University of Guelph, Guelph, ON (contributed poster presentation)
- Angevaare, J.✦ (2014) Fitting disease models with likelihoods. Community of Interest in Disease Modelling, University of Guelph, Guelph, ON (oral presentation)
- Angevaare, J.✦, Gillis, D., Darlington, G. (2014) Efficient Bayesian Inference for Conditionally Autoregressive Models. Annual Meeting of the Statistical Society of Canada, University of Toronto, Toronto, ON (contributed poster presentation)
- Angevaare, J.✦, Gillis, D. (2012). The Utility of Catch Per Unit Effort Variance. Annual Meeting of the Statistical Society of Canada, University of Guelph, Guelph, ON (contributed poster presentation)
- Gillis, D.✦, Angevaare, J. (2012). Something’s Fishy. Colloquium Joint BIOM&S Seminar Series, University of Guelph, Guelph, ON (invited presentation)
Theses
- Angevaare, J. (2020) Phylodynamic and Transmission Network Individual Level Infectious Disease Models. Department of Mathematics and Statistics, University of Guelph, Guelph, ON. Doctoral Thesis. Sept 2020. 141 pp. Advisors: Prof. Zeny Feng, Prof. Rob Deardon.
- Angevaare, J. (2014) Efficient Bayesian Inference for Conditionally Autoregressive Models. Department of Mathematics and Statistics, University of Guelph, Guelph, ON. Master's Thesis. April 2014. 122 pp. Advisors: Prof. Daniel Gillis, Prof. Gerarda Darlington.
Technical documents
- Gillis, D., Alexander, C., Angevaare, J., Bakar, C., Cox, R., Kramski, N., Krsic, N. (2014) SON-BP Collaborative Lake Whitefish Research Project: Year 3 report. Technical document.
- Angevaare, J., Gillis, D., Cox, R. (2014) SON-BP Collaborative Lake Whitefish Research Project: Milestone 5: Report on Application and Evaluation of Population Models. Technical document. June 2014. 28 pp.
- Angevaare, J., Gillis, D. (2014) SON-BP Collaborative Lake Whitefish Research Project: Milestones C3 & C4 - Estimates of Life History and Mortality Parameters of Lake Whitefish (Coregonus clupeaformis). Technical document. January 2014. 15 pp.
- Gillis, D., Angevaare, J., Rueffer, M., Horrocks, J. (2012). Analysis of Total Allowable Catch. Technical document. October 2012. 34 pp.
- Angevaare, J. (2012) Lake Huron Lake Whitefish Life History Parameters. Technical document. September 2012. 106 pp. (including appendices)
Previous software projects
FishABM.jl (2014-2015)FishABM.jl was an implementation of a lifecycle model for the application of fisheries management. An age-structured model was used for adults, with options for commercial or recreational harvest. A highly detailed agent based model was used from egg through to juvenilles life stages, grouping individuals from the same brood into an single agent. Movement and various sources of mortality (habitat specific), and the eventual impact on assumptions surrounding movement and mortality on the health of the fishery could be tracked.
Fisheries_ABM (2014)Fisheries_ABM was a prototype for would eventually become FishABM.jl, built in R. It was an stochastic agent based model of a fishery that could account for harvest and anthropogenic mortality sources.
ILMs.jl (2014-2015)ILMs.jl was my first implementation of individual level models of infectious disease in Julia. It offered simulation and inference in continuous and discrete time, when event times can assumed to be known, for models utilizing power-law infectivity kernel. This package is superceded by my more recent and involved work with Pathogen.jl. My experience developing this package convinced me to go all-in with Julia for my PhD research.
ILMPythonTools (2014)ILMPythonTools was an early python implementation of tools for individual level models of infectious disease. This work was stopped in favour of developing such tools instead in Julia.
Just for fun
brew2 (2018-2022)brew2 is an app built in Node-RED for brewery control using Raspberry Pi. It implements sophisticated PID algorithms for mash temperature control using various recirculating mash systems. Its more custom features are written in javascript. Logging uses influxdb time series database.

I have developed several popular plugins for CraftBeerPi3 brewery control software in python:
HopScrape (2018)This project incorporated scraping, and imputation of hop data using BeautifulSoup, pandas, and fancyimpute. The cleaned and imputed data resulting from this project are available on the repo.
TriangleDash (2018)TriangleDash is a small application I made to learn Plotly's Dash. It is an explanation, calculation, and visualization of results from triangle test sensory analysis.
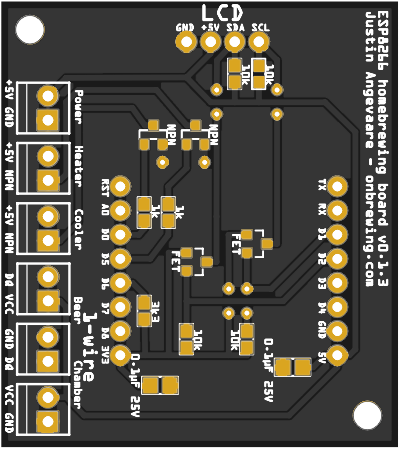
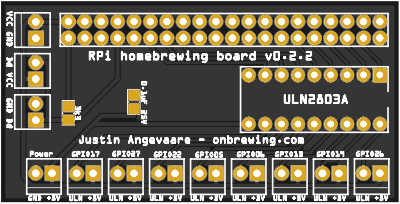
Homebrewing PCBs (2018-2020)I learned a bit about PCB design and electronics, then created a couple board designs for homebrewing applications using Raspberry Pi and ESP8266 IoT devices and open-sourced them. They can also be purchased from PCBWay.com1, 2. 100s of these boards have now been produced.